The Repsc R package is a beta development version to facilitate expression analysis of transposable elements at single-cell resolution.
Functionality
The basic workflow consists importing genome annotation/sequencing reads, computing the count matrix, and QC of the results.
Features include:
- Multi-species support
- Flexible genome annotation
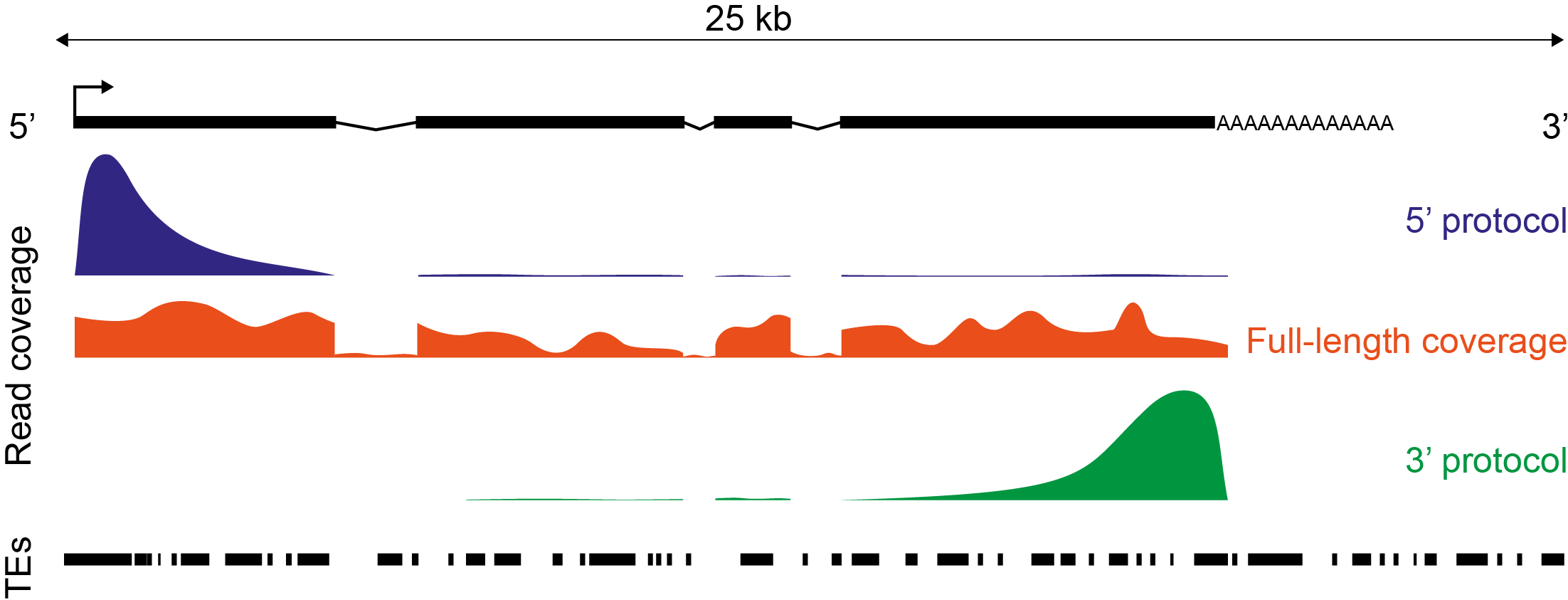
- 5’/3’/full-coverage support
- High-quality reports
More details on the usage of Repsc is available in the package vignette.
Installation
# Install BiocManager (in case you haven't already)
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
# Install Repsc
install.packages('Repsc', repos = 'tanaylab.bitbucket.io/repo')Note: Repsc requires R version 3.5 or higher. The package was tested on linux and macbook machines, but is currently not compatible on Windows. A typical application will require at least 4 GB RAM, but heavier use cases (e.g. large BAM files) may require significantly more. For improved speed performance, we recommend a multi-CPU/core workstation.
