Plot LOGO of the pssm result from the regression
plot_pssm_logo.RdPlot LOGO of the pssm result from the regression
Usage
plot_pssm_logo(
pssm,
title = "Sequence model",
subtitle = ggplot2::waiver(),
pos_bits_thresh = NULL,
revcomp = FALSE,
method = "bits"
)Arguments
- pssm
PSSM matrix or data frame
- title
title of the plot
- subtitle
subtitle of the plot
- pos_bits_thresh
Positions with bits above this threshold would be highlighted in red. If
NULL, no positions would be highlighted.- revcomp
whether to plot the reverse complement of the PSSM
- method
Height method, can be one of "bits" or "probability" (default:"bits")
Examples
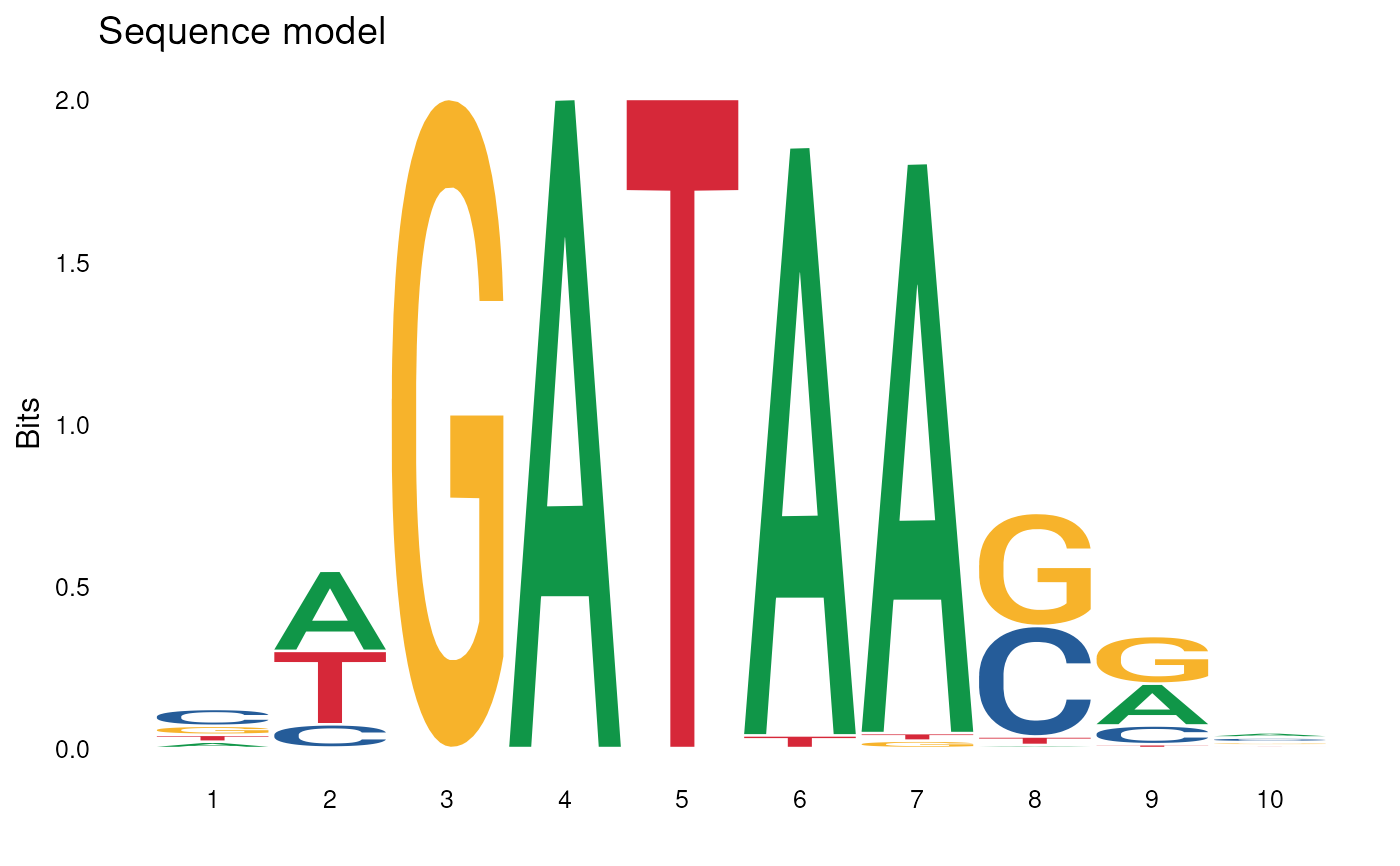
pssm <- data.frame(
pos = seq(0, 9, by = 1),
A = c(
0.16252439817826936, 0.4519127838188067, 0, 1, 0, 0.9789171974522293,
0.9743866100297978, 0.013113942843003835, 0.3734676916683981,
0.32658771473191045
),
C = c(
0.43038386467143785, 0.13116231900388756, 0, 0, 0, 0, 0, 0.46975132995175056,
0.1669956368169541, 0.29795679333680375
),
G = c(
0.22999349381912818, 0.002929742520705392, 1, 0, 0, 0, 0.012679896024852597,
0.4808858097241123, 0.4248389777685435, 0.20458094742321709
),
T = c(
0.1770982433311646, 0.41399515465660036, 0, 0, 1, 0.0210828025477707,
0.012933493945349648, 0.03624891748113324, 0.0346976937461043,
0.17087454450806872
)
)
plot_pssm_logo(pssm)
 if (FALSE) { # \dontrun{
res <- regress_pwm(sequences_example, response_mat_example)
plot_pssm_logo(res$pssm)
} # }
if (FALSE) { # \dontrun{
res <- regress_pwm(sequences_example, response_mat_example)
plot_pssm_logo(res$pssm)
} # }