10 QC - sc methylation
10.0.1 initialize definitions
## cgdb object
## 21,342,746 CpGs X 24,179 cells## --- root (@db_root): /net/mraid14/export/tgdata/users/aviezerl/proj/ebdnmt/Dnmt3ab_EB/methylation/data/cgdbsc_stats <- sc_stats %>%
left_join(invivo_sort) %>%
replace_na(replace = list(germ_layer = "other")) %>%
mutate(sort = ifelse(day == "e7.5", germ_layer, sort)) ## Joining, by = "cell_id"Note that for cell cycle we use specific experiments with similar cell cycle profile
## # A tibble: 15 x 4
## day line sort n
## 1 d5 ko3a CXCR4-EPCAM+ 372
## 2 d5 ko3a CXCR4+EPCAM+ 372
## 3 d5 ko3a index 1851
## 4 d5 ko3b CXCR4-EPCAM+ 372
## 5 d5 ko3b CXCR4+EPCAM+ 372
## 6 d5 ko3b index 1837
## # ... with 9 more rows10.2 Number of cells
10.2.1 Extended Data Figure 9A
## # A tibble: 4 x 4
## samp_id day line n
## 1 d5_3a d5 ko3a 2217
## 2 d5_3b d5 ko3b 2197
## 3 d5_wt d5 wt 2984
## 4 e7.5 e7.5 mouse 2146## # A tibble: 2 x 2
## day n
## 1 d5 7398
## 2 e7.5 2146p <- sc_stats %>%
filter(cg_num >= 2e4) %>%
organize_sc_samp_id() %>%
count(samp_id, day, line) %>%
mutate(samp_id = factor(samp_id, levels=samp_levels)) %>%
ggplot(aes(x=samp_id, y=n)) +
geom_col() +
vertical_labs() +
ylab("# of cells with more than 20k CpGs") +
xlab("")
p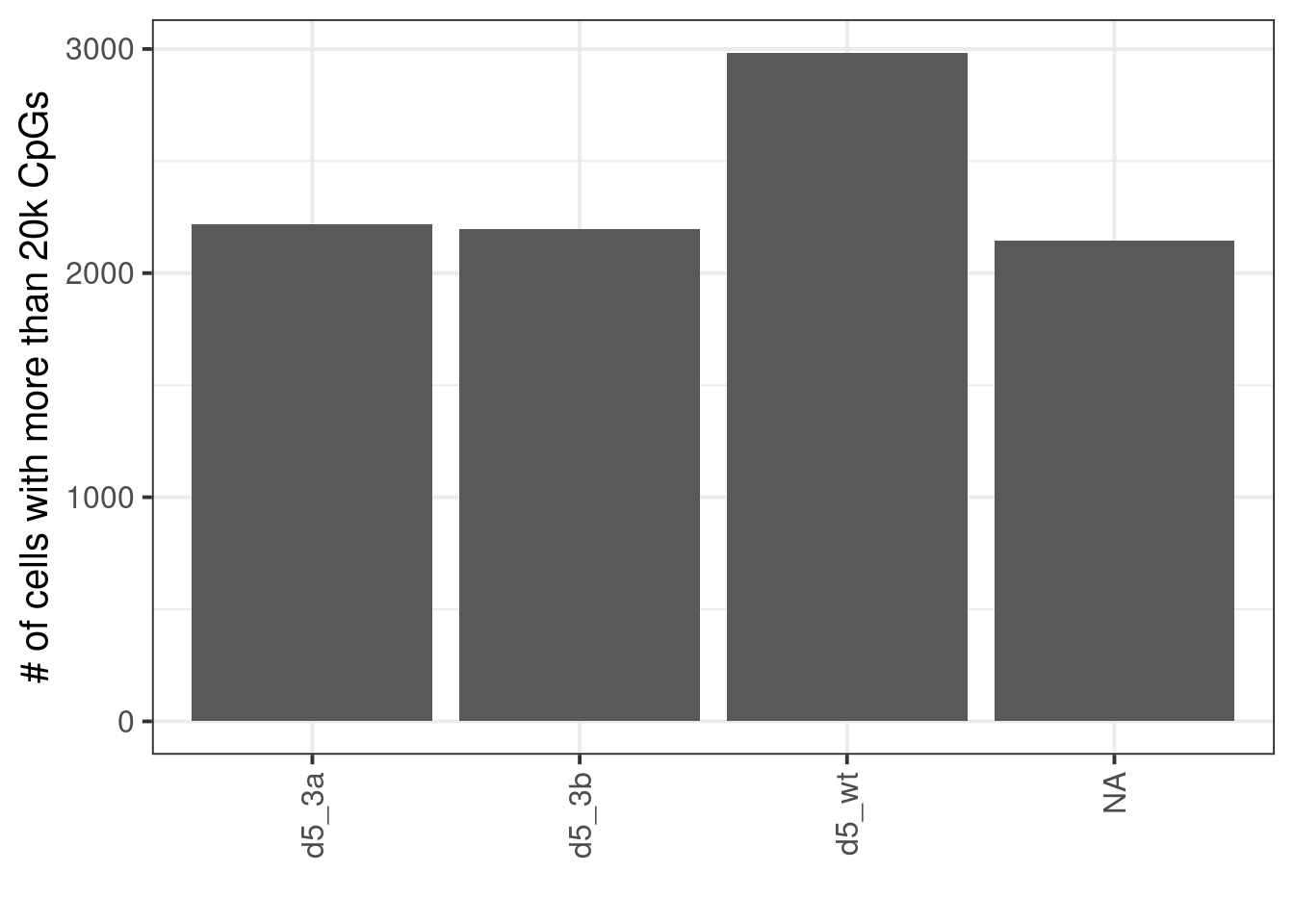
10.3 Number of CpGs
10.3.1 Extended Data Figure 9C
p <- sc_stats %>%
filter(cg_num >= 2e4) %>%
organize_sc_samp_id() %>%
mutate(samp_id = factor(samp_id, levels=samp_levels)) %>%
ggplot(aes(x=samp_id, y=cg_num)) +
geom_boxplot(fill="gray", outlier.shape = NA) +
scale_y_log10(labels=scales::scientific) +
ggforce::geom_sina(size=0.001, alpha=0.01) +
vertical_labs() +
ylab("# of CpGs") +
xlab("")
p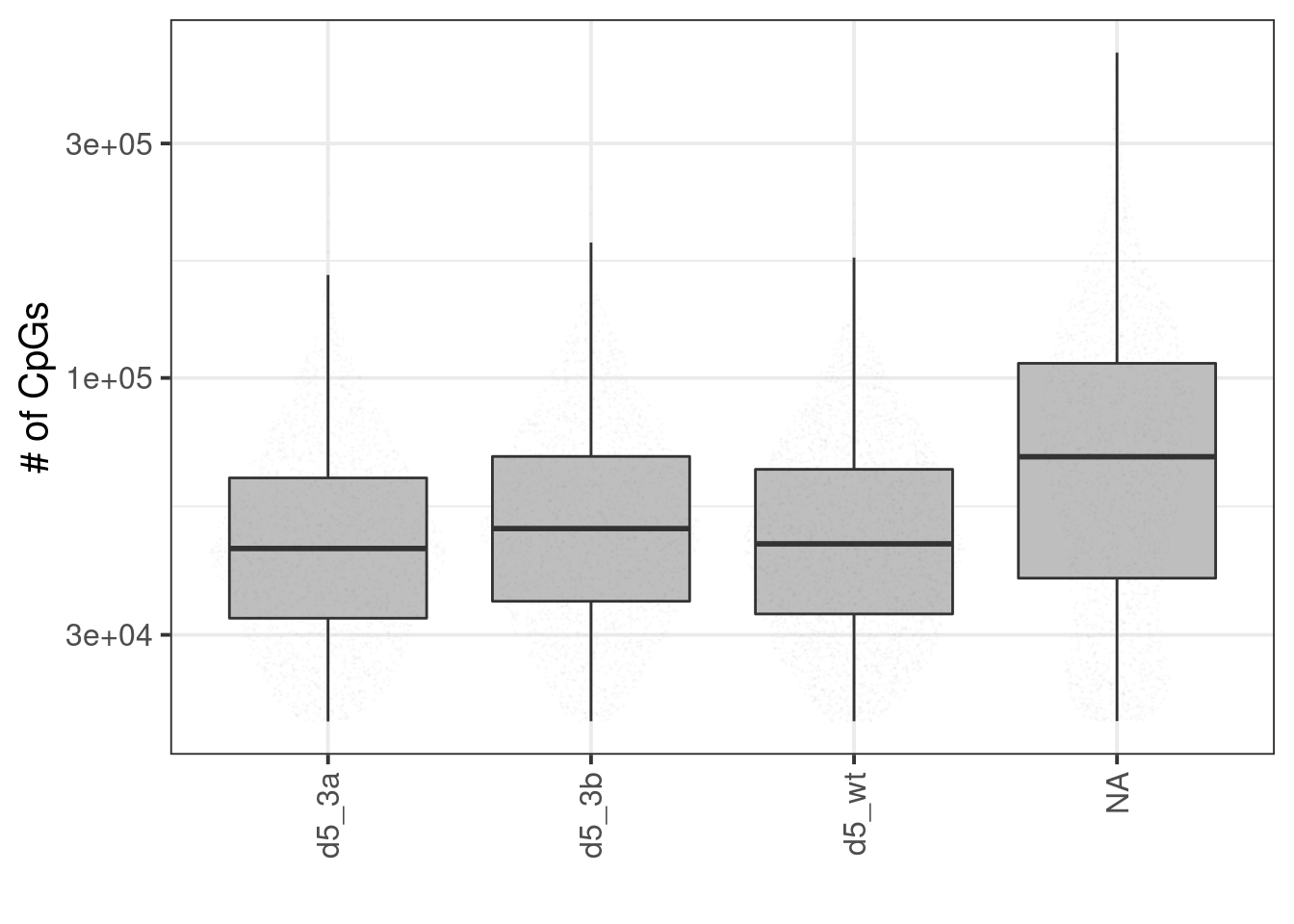
10.4 Sorting plots
facs_df <- sc_stats %>%
filter(!is.na(APC_A), !is.na(BV421_A)) %>%
group_by(sort_date) %>%
mutate(EPCAM = log2(abs(min(APC_A, na.rm=TRUE)) + 10 + APC_A), CXCR4 = log2(abs(min(BV421_A, na.rm=TRUE)) + 10 + BV421_A)) %>%
replace_na(replace = list(gate = "no_gate")) %>%
ungroup() %>%
filter(cpg_num >= 2e4)
facs_df_e7.5 <- facs_df %>% filter(day == "e7.5")
facs_df_d5 <- facs_df %>% filter(day == "d5")10.5 Early/Late coverage per cell (e7.5)
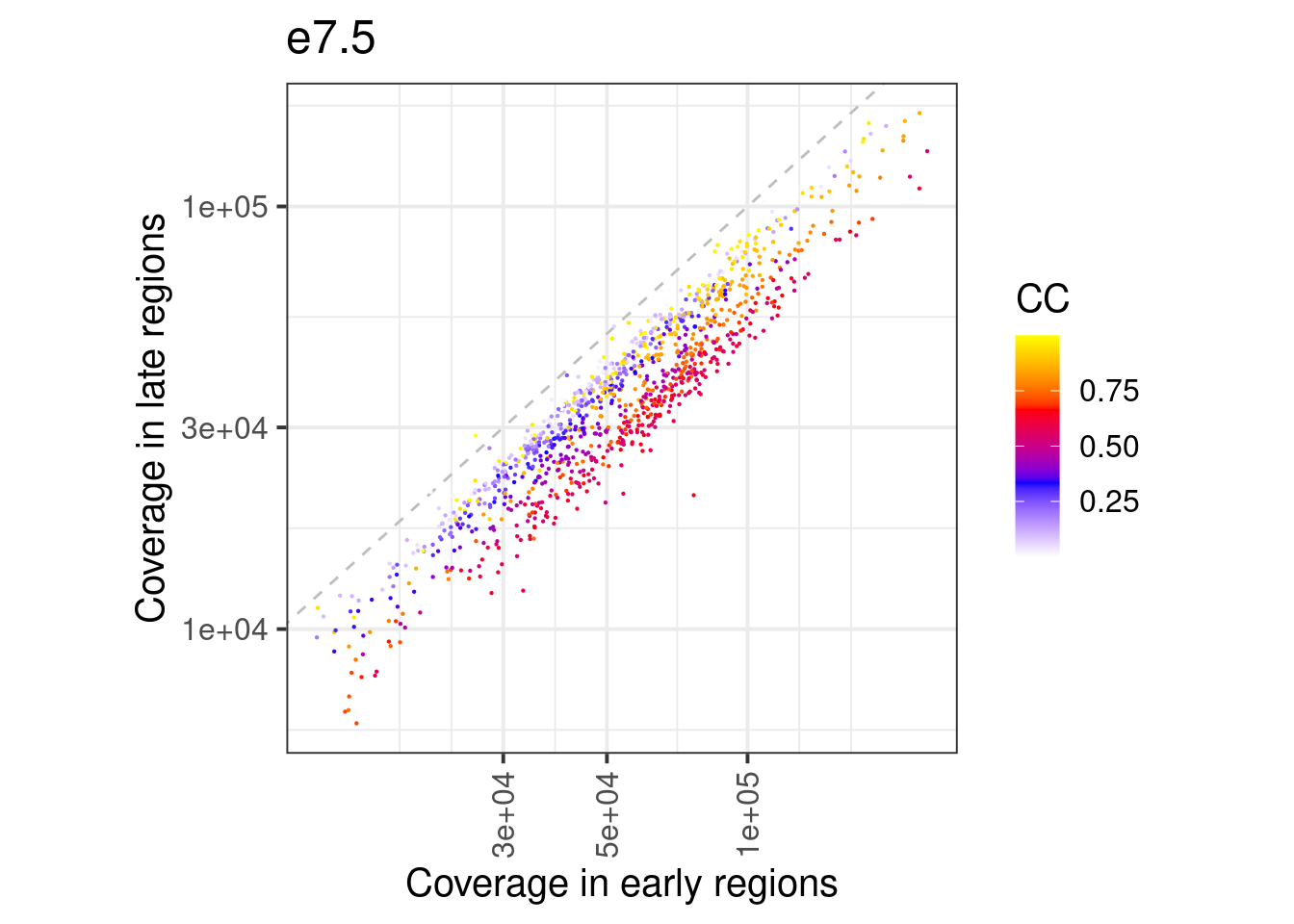
df_ord_invivo <- fread(here("output/cell_cycle/segmented.tsv")) %>% filter(day == "e7.5") %>% mutate(germ_layer = factor(line, levels=c("ecto", "meso", "endo")))10.5.1 Extended Data Figure 9F
p <- df_ord_invivo %>%
ggplot(aes(x=early, y=late, color=ord2)) +
geom_abline(linetype="dashed", color="gray") +
geom_point(size=0.1) +
scale_color_gradientn(name = "CC", colors=c("white", "blue", "red", "yellow")) +
theme(aspect.ratio=1) +
scale_x_log10(labels=scales::scientific) +
scale_y_log10(labels=scales::scientific) +
vertical_labs() +
xlab("Coverage in early regions") +
ylab("Coverage in late regions") +
ggtitle("e7.5")
p