6 Yagi et al.
6.0.2 DMRs
yagi_dmrs <- fread(here("data/yagi_dmrs.tsv")) %>%
filter(chrom %in% gintervals.all()$chrom) %>%
as_tibble()
yagi_dmrs <- yagi_dmrs %>%
mutate(
Yagi_3a = ifelse(type == "proA", meanMethy2, meanMethy1),
Yagi_3b = ifelse(type == "proB", meanMethy2, meanMethy1)
)
yagi_dmrs <- yagi_dmrs %>%
select(chrom, start, end, type, Yagi_3a, Yagi_3b)
head(yagi_dmrs)## # A tibble: 6 x 6
## chrom start end type Yagi_3a Yagi_3b
## 1 chr6 48387929 48389217 proA 0.05732420 0.6731291
## 2 chr3 55584536 55588340 proA 0.02412445 0.4470477
## 3 chr1 186549516 186554917 proA 0.02829836 0.5260327
## 4 chr4 135845931 135846972 proA 0.10997219 0.6153078
## 5 chr11 118941147 118943355 proA 0.04548308 0.6741248
## 6 chr7 77491664 77494580 proA 0.02552988 0.6597258## [1] 7646.0.3 Get Meissner data
md <- tribble(~track_name, ~sort, ~num,
"Meissner_Nature_2017.Epi_Dnmt3a_1", "ko3a", 1,
"Meissner_Nature_2017.Epi_Dnmt3a_2", "ko3a", 2,
"Meissner_Nature_2017.Epi_Dnmt3a_4", "ko3a", 4,
"Meissner_Nature_2017.Epi_Dnmt3a_5", "ko3a", 5,
"Meissner_Nature_2017.Epi_Dnmt3a_6", "ko3a", 6,
"Meissner_Nature_2017.Epi_Dnmt3b_1", "ko3b", 1,
"Meissner_Nature_2017.Epi_Dnmt3b_2", "ko3b", 2,
"Meissner_Nature_2017.Epi_Dnmt3b_3", "ko3b", 3,
"Meissner_Nature_2017.Epi_Dnmt3b_4", "ko3b", 4,
"Meissner_Nature_2017.Epi_Dnmt3b_5", "ko3b", 5,
"Meissner_Nature_2017.Epi_Dnmt3b_6", "ko3b", 6,
"Meissner_Nature_2017.Epi_WT_4", "wt", 4,
"Meissner_Nature_2017.Epi_WT_5", "wt", 5,
"Meissner_Nature_2017.Epi_WT_6", "wt", 6,
"Meissner_Nature_2017.Epi_WT_7", "wt", 7) %>%
mutate(name = paste0(sort, "_", num))
m_all_epi6 <- gextract_meth(tracks = md$track_name, names=md$name, intervals=yagi_dmrs %>% select(chrom, start, end), extract_meth_calls = TRUE, iterator = yagi_dmrs %>% select(chrom, start, end)) %cache_df% here("output/meissner_epi_yagi_meth.tsv") %>% as_tibble() min_cov <- 10
m_epi6 <- m_all_epi6 %>% select(chrom, start, end)
for (g in unique(md$sort)){
nms <- md %>% filter(sort == g) %>% pull(name)
cov_col <- paste0(g, ".cov")
meth_col <- paste0(g, ".meth")
m_epi6[[cov_col]] <- rowSums(m_all_epi6[, paste0(nms, ".cov")], na.rm=TRUE)
m_epi6[[meth_col]] <- rowSums(m_all_epi6[, paste0(nms, ".meth")], na.rm=TRUE)
m_epi6[[g]] <- ifelse(m_epi6[[cov_col]] >= min_cov, m_epi6[[meth_col]] / m_epi6[[cov_col]], NA)
}
m_epi6 <- m_epi6 %>% select(-ends_with(".meth"), -ends_with(".cov")) %cache_df% here("output/meissner_epi_yagi_meth_sum.tsv")6.0.4 Get MEEB data
m_meeb <- calc_eb_day0_to_day6_cpg_meth(intervals = yagi_dmrs %>% select(chrom, start, end), iterator = yagi_dmrs %>% select(chrom, start, end), cache_fn = here("output/meeb_day0_to_day6_yagi_meth.tsv"), use_sort = FALSE, max_na = 100) %>%
select(-ends_with("cov"), -ends_with("meth")) %>%
select(-contains("_ko1"), -contains("_tko"), -starts_with("d0_"))## # A tibble: 6 x 30
## chrom start end d3_d13ako d4_d13ako d3_d13bko d4_d13bko d5_dko
## 1 chr1 14497410 14498475 NA NA NA 0.38461538 NA
## 2 chr1 21070410 21070719 NA NA NA NA NA
## 3 chr1 34163709 34164326 NA NA NA NA NA
## 4 chr1 38266943 38267649 0.1076265 0.09757137 0.1166078 0.07476636 0.01094891
## 5 chr1 38685382 38686284 NA NA NA NA NA
## 6 chr1 57032568 57032760 0.0000000 0.00000000 0.0000000 NA NA
## d6_dko d0S_3a d1_3a d2_3a d3_3a d4_3a d5_3a
## 1 NA 0.66346154 NA NA NA 0.55855856 0.4925373
## 2 NA 0.31428571 NA NA NA 0.29113924 0.3614458
## 3 NA 0.05769231 NA NA NA 0.08333333 0.1320755
## 4 0.009803922 0.29095853 0.1721992 0.1582915 0.2002882 0.25531915 0.5327103
## 5 NA 0.41111111 NA NA NA 0.22222222 0.4615385
## 6 NA 0.06779661 NA NA 0.0000000 0.00000000 0.0000000
## d6_3a d0S_3b d1_3b d2_3b d3_3b d4_3b d5_3b
## 1 0.50000000 0.8068182 NA NA NA 0.80373832 0.80000000
## 2 0.43801653 0.5918367 NA NA NA 0.61538462 0.64406780
## 3 0.17283951 0.0000000 NA NA NA 0.13513514 0.22580645
## 4 0.67054908 0.3020355 0.1372951 0.2084993 0.2549451 0.25453654 0.29600000
## 5 0.44520548 0.5306122 NA NA NA 0.48913043 0.64444444
## 6 0.04635762 0.1538462 NA NA NA 0.04395604 0.01923077
## d6_3b d0S_wt d1_wt d2_wt d3_wt d4_wt d5_wt
## 1 0.81250000 NA NA NA 0.8571429 0.8000000 0.79569892
## 2 0.67832168 NA NA NA NA 0.9523810 0.59183673
## 3 0.22602740 NA NA NA NA 0.1600000 0.46913580
## 4 0.33012628 0.3144775 0.1382979 0.1384615 0.2685560 0.3562982 0.77044610
## 5 0.58235294 NA NA NA NA 0.5666667 0.66666667
## 6 0.05128205 NA NA NA NA 0.2666667 0.03921569
## d6_wt
## 1 0.82173913
## 2 0.84771574
## 3 0.36200000
## 4 0.74321230
## 5 0.61437908
## 6 0.05809129## [1] 764 30## Joining, by = c("chrom", "start", "end")
## Joining, by = c("chrom", "start", "end")## # A tibble: 6 x 36
## chrom start end type Yagi_3a Yagi_3b ko3a ko3b
## 1 chr6 48387929 48389217 proA 0.05732420 0.6731291 0.03305727 0.01935100
## 2 chr3 55584536 55588340 proA 0.02412445 0.4470477 0.03345408 0.02190160
## 3 chr1 186549516 186554917 proA 0.02829836 0.5260327 0.03760639 0.02147777
## 4 chr4 135845931 135846972 proA 0.10997219 0.6153078 0.11737341 0.04451465
## 5 chr11 118941147 118943355 proA 0.04548308 0.6741248 0.07684360 0.03943182
## 6 chr7 77491664 77494580 proA 0.02552988 0.6597258 0.00000000 0.04000000
## wt d3_d13ako d4_d13ako d3_d13bko d4_d13bko d5_dko d6_dko
## 1 0.03128703 NA 0.05000000 NA 0.00000000 NA 0.00000000
## 2 0.05538695 NA 0.00000000 0.0000000 0.00000000 NA NA
## 3 0.07845847 0.09322034 0.09478407 0.1114696 0.04453912 0.007029877 0.01030928
## 4 0.21266968 0.03361345 0.06903766 0.0311804 0.02857143 0.000000000 0.01162791
## 5 0.14754879 0.15384615 0.09090909 0.0000000 0.03571429 NA 0.00000000
## 6 NA NA 0.00000000 NA NA NA NA
## d0S_3a d1_3a d2_3a d3_3a d4_3a d5_3a d6_3a
## 1 0.05240642 0.00000000 0.0000000 0.03378378 0.03199269 0.0715859 0.1162437
## 2 0.22588832 0.31034483 0.1200000 0.08333333 0.24896836 0.2323718 0.2582781
## 3 0.46839605 0.33613445 0.3232462 0.38888889 0.43388339 0.2618705 0.3223706
## 4 0.12714157 0.06666667 0.0505618 0.10747664 0.10019084 0.1414427 0.4215470
## 5 0.18901660 0.07692308 0.2500000 0.14893617 0.19910847 0.2071713 0.2143895
## 6 0.36474164 0.25000000 0.1153846 0.34482759 0.34335840 0.2874251 0.3011665
## d0S_3b d1_3b d2_3b d3_3b d4_3b d5_3b d6_3b
## 1 0.09992194 0.01086957 0.02816901 0.00781250 0.03423237 0.05020921 0.03491379
## 2 0.22635659 0.19148936 0.12500000 0.14893617 0.21798030 0.21082621 0.25292056
## 3 0.51350450 0.40802213 0.51413428 0.60601504 0.54364798 0.23013245 0.35954601
## 4 0.11684518 0.03960396 0.05479452 0.09459459 0.10903427 0.08480000 0.08768536
## 5 0.19321149 0.23255814 0.14285714 0.22580645 0.18750000 0.17412141 0.18470021
## 6 0.66250000 0.15384615 0.88235294 0.77777778 0.72745098 0.64102564 0.62837163
## d0S_wt d1_wt d2_wt d3_wt d4_wt d5_wt d6_wt
## 1 0.0733945 0.00000000 0.01075269 0.04411765 0.05958132 0.07562278 0.08210877
## 2 0.1935484 0.11363636 0.18918919 0.22857143 0.27392739 0.31428571 0.33866951
## 3 0.6337895 0.39802336 0.44246032 0.56379101 0.67888716 0.59645937 0.38639532
## 4 0.1174785 0.05925926 0.05384615 0.12921348 0.19919786 0.27834179 0.32883390
## 5 0.1296296 0.06000000 0.07317073 0.15789474 0.28443114 0.22222222 0.24428214
## 6 0.7187500 0.48275862 0.66666667 0.56410256 0.82432432 0.73489933 0.66535433## # A tibble: 2 x 2
## type n
## 1 proA 427
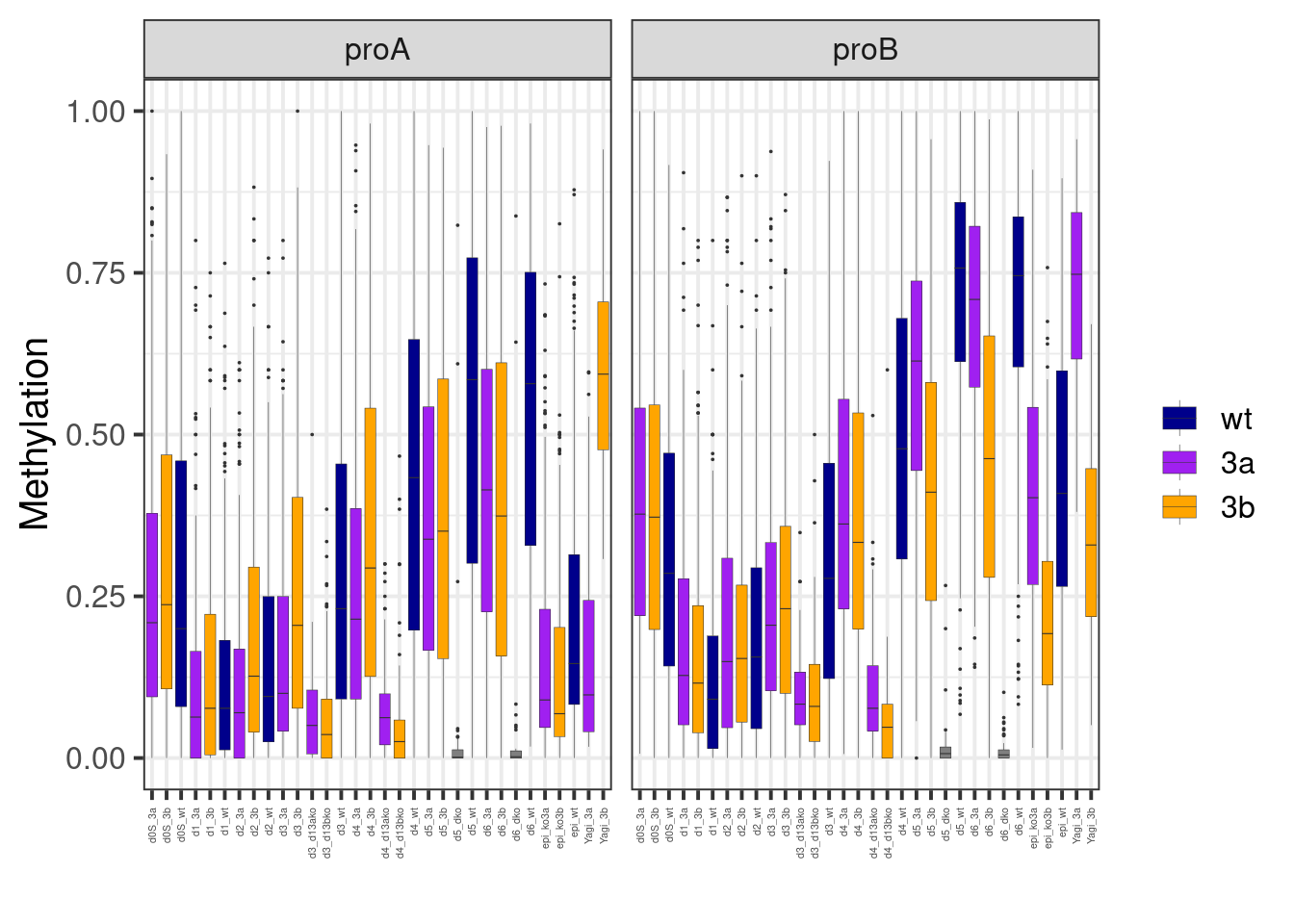
## 2 proB 3436.0.5 Extended Data Figure 8I
options(repr.plot.width = 14, repr.plot.height = 7)
line_colors <- c("wt" = "darkblue", "3a" = "purple", "3b" = "orange")
p_yagi <- yagi_meth %>%
select(chrom:end, type, Yagi_3a, Yagi_3b, epi_ko3a = ko3a, epi_ko3b = ko3b, epi_wt = wt, everything()) %>%
gather("samp", "meth", -(chrom:type)) %>%
mutate(samp = gsub("_all", "", samp)) %>%
mutate(line = case_when(
grepl("3a", samp) ~ "3a",
grepl("3b", samp) ~ "3b",
grepl("wt", samp) ~ "wt"
)) %>%
mutate(line = factor(line, levels = names(line_colors))) %>%
ggplot(aes(x=samp, y=meth, fill=line)) +
geom_boxplot(outlier.size = 0, lwd = 0.1) +
xlab("") +
ylab("Methylation") +
scale_fill_manual("", values=line_colors) +
vertical_labs() +
theme(axis.text.x=element_text(size=4)) +
facet_wrap(~type)
p_yagi## Warning: Removed 8158 rows containing non-finite values (stat_boxplot).