Supervised filtering of feature genes: guided tutorial on 8K PBMCs
Yaniv Lubling
2018-10-10
Source:vignettes/b-geneset_by_anchor_pbmc8k.Rmd
b-geneset_by_anchor_pbmc8k.RmdIntroduction
This note walks through the common scenario of disabling specific gene modules from affecting the partitioning of cells into metacells. In many applications we would want to mask technical effects of stress (e.g. heat shock) from affecting the partitioning, or to mask cell cycle related genes in order to get metacells that represent cell types/states and not stages at the cell cycle.
We'll start with finding genes that are correlated to a few cell cycle and stress halmark genes, then generate gene correlation heatmaps that will help to further filter the genes, and lastly show how to create metacells that are not affected by these genes.
We use here the small 8K PBMC dataset that is used in the Running metacell analysis: guided tutorial on 8K PBMCs vignette, please make sure to run that vignette before executing this one, since we'll use several objects produced there.
Setting up
We start by loading the library and initializating the database and the figures directories:
library("metacell")
scdb_init("testdb/", force_reinit=T)
#> initializing scdb to testdb/
figs_dir = "figs"
scfigs_init(figs_dir)Creating the gene correlation matrix
We're going to generate a gene-gene correlation matrix using the umi matrix generated in the Running metacell analysis: guided tutorial on 8K PBMCs vignette. The genes will then be filtered by thresholding their correlation with the user supplied genes (termed anchor genes). In this vignette we'll build a set of cell cycle and stress related genes. So let's define these gene anchors, and some additional variables:
genes_anchors = c('PCNA', 'TOP2A', 'TXN', 'HSP90AB1', 'FOS')
tab_fn = paste(figs_dir, "lateral_gmods.txt", sep="/")
gset_nm = "lateral"tab_fn is the output file name of the correlation matrix, and gset_nm is the id of the gene set that will be generated.
Now we'll call the function that computes the gene-gene correlation matrix:
mcell_mat_rpt_cor_anchors(mat_id="test", gene_anchors = genes_anchors, cor_thresh = 0.1, gene_anti = c(), tab_fn = tab_fn, sz_cor_thresh = 0.2)The function computes the correlations of all genes in the umi matrix with the gene anchors after downsampling the umi counts. It then filters genes that their correlation to the anchor genes is above cor_thresh. Genes can also be filtered based on their correlation to a set of negative control genes, that can be specified in the gene_anti parameter. In this case, genes that their top correlated gene is a negative control and not one of the anchor genes are discarded. If the sz_cor_thresh is not NA, genes with correlation to the cells total umi counts above sz_cor_thresh are also selected. In our case, we expect some cell cycle genes to be correlated with cell size, so we specify a positive threshold.
Let's read the filtered gene correlation matrix and examine it:
gcor_mat = read.table(tab_fn, header=T)
head(gcor_mat)
#> sz_cor PCNA TOP2A TXN HSP90AB1
#> AC023590.1 0.046756633 0.010617491 -0.003780935 0.18146371 -0.00416468
#> ACTB 0.250543976 0.059445816 0.111477842 0.13386438 -0.05907979
#> ACTG1 0.213020480 0.045909657 0.094956945 0.04625107 0.04491417
#> AGTRAP 0.001536675 0.010654896 -0.009926729 0.07963326 -0.07378579
#> AHNAK 0.057235736 -0.005677824 0.002048814 0.09009918 -0.04284507
#> AHR 0.054036998 -0.005936477 -0.007005925 0.05127561 -0.04556298
#> FOS max neg_max
#> AC023590.1 -0.01856910 0.18146371 0
#> ACTB 0.13500093 0.13500093 0
#> ACTG1 -0.08386522 0.09495695 0
#> AGTRAP 0.14986172 0.14986172 0
#> AHNAK 0.13974787 0.13974787 0
#> AHR 0.10690160 0.10690160 0
print(dim(gcor_mat))
#> [1] 560 8The row names are genes, and the columns are the correlation to the total umi counts (sz_cor), the correlations to the anchor genes and their maximal one (max). Since we didn't specify negative anchor genes in this example the maximal correlation to the negative anchors is zero (max_neg).
Now we'll add a gene set to the database, comprised of the genes in the filtered matrix:
foc_genes = apply(gcor_mat[, genes_anchors], 1, which.max)
gset = gset_new_gset(sets = foc_genes, desc = "Cell cycle and stress correlated genes")
print(gset)
#> An object of class "tgGeneSets"
#> Slot "description":
#> [1] "Cell cycle and stress correlated genes"
#>
#> Slot "set_names":
#> [1] 3 5 2 4 1
#>
#> Slot "gene_set":
#> AC023590.1 ACTB ACTG1 AGTRAP AHNAK
#> 3 5 2 5 5
#> AHR AIF1 ALDH1A1 ALDH2 ALDH3B1
#> 5 5 5 5 5
#> ALDOA ALOX5AP ANKRD28 ANPEP ANXA1
#> 5 3 5 5 5
#> ANXA2 ANXA5 AOAH AP1S2 AP2S1
#> 3 5 5 5 5
#> APLP2 APP ARHGEF40 ARL5B ARL6IP1P2
#> 5 3 5 5 2
#> ARRDC4 ASAH1 ASF1B ASGR1 ASIP
#> 5 5 2 5 3
#> ATF3 ATG3 ATOX1 ATP2B1-AS1 ATP5E
#> 5 5 5 5 5
#> ATP5G2 ATP5I ATP6V0B ATP6V0D1 ATP6V0E1
#> 5 5 5 5 5
#> ATP6V1A ATP6V1B2 ATP6V1F BACH1 BCAT1
#> 5 5 5 5 3
#> BCL2A1 BIRC5 BLOC1S1 BLVRB BNIP3L
#> 5 2 5 5 5
#> BRI3 BST1 C10orf11 C12orf75 C14orf2
#> 5 5 5 3 5
#> C19orf38 C19orf47 C1orf162 C20orf27 C4orf48
#> 5 2 5 5 5
#> C5AR1 C7orf73 C9orf72 CALM2 CAMK1
#> 5 5 5 5 5
#> CAPG CAPZA2 CARD16 CARS2 CASP1
#> 5 5 5 5 5
#> CATSPER1 CCDC189 CCDC50 CCDC71L CCDC88A
#> 5 3 3 2 5
#> CCL3 CCL3L3 CCNA2 CCNL1 CCNY
#> 5 5 2 5 5
#> CCR1 CCR9 CD14 CD163 CD300E
#> 5 2 5 5 5
#> CD300LF CD302 CD33 CD36 CD4
#> 5 5 5 5 5
#> CD44 CD63 CD70 CD83 CD86
#> 5 5 2 5 5
#> CD93 CDA CDC42 CDCA8 CDK1
#> 5 5 5 2 2
#> CDK2 CDKN2C CDKN3 CDR2 CEBPB
#> 2 2 2 2 5
#> CEBPD CECR1 CENPE CENPF CENPN
#> 5 5 2 2 2
#> CENPP CENPU CFD CFP CHMP1B
#> 2 2 5 5 5
#> CHP1 CIB2 CKAP4 CKLF CLEC10A
#> 5 3 5 5 3
#> CLEC12A CLEC4A CLEC4C CLEC4E CLEC7A
#> 5 5 3 5 5
#> CMTM6 CNPY3 COMT CORO1A COTL1
#> 5 5 3 2 5
#> COX6B1 COX7B CPPED1 CPVL CREB5
#> 5 5 5 5 5
#> CREG1 CRTAP CRYM CSF3R CST3
#> 5 5 3 5 5
#> CSTA CTB-61M7.2 CTC-510F12.4 CTD-3252C9.4 CTSB
#> 5 5 5 5 5
#> CTSC CTSD CTSH CTSS CTSZ
#> 3 5 5 5 5
#> CXCL2 CXCL8 CYBA CYBB CYP1B1
#> 5 5 5 5 5
#> DAZAP2 DERL3 DMXL2 DNASE1L3 DOK3
#> 5 3 5 3 5
#> DPM2 DPYD DPYSL2 DSTNP2 DUSP1
#> 2 5 5 2 5
#> DUSP6 DYNLL1 EFCAB11 EGR1 EHBP1L1
#> 5 3 2 5 5
#> EIF1 EIF4EBP1 ELOB ENHO ENTPD1
#> 5 5 5 3 5
#> EREG EVI2A EVI2B FAM120A FAM198B
#> 5 5 5 5 5
#> FAM45A FANCI FCAR FCER1A FCER1G
#> 5 2 5 3 5
#> FCGR1A FCGR2A FCGRT FCN1 FES
#> 5 5 5 5 5
#> FGD4 FGL2 FKBP1A FKBP2 FLT3
#> 5 5 5 3 3
#> FOLR3 FOS FOSB FOSL2 FPR1
#> 5 5 5 5 5
#> FTH1 FTL FTLP3 G0S2 GABARAPL1
#> 5 5 5 5 5
#> GAPDH GAS6 GAS7 GCA GLIPR1
#> 5 3 5 5 5
#> GLRX GLUL GMFG GNAQ GNS
#> 5 5 5 5 5
#> GPBAR1 GPCPD1 GPX1 GRINA GRN
#> 5 5 5 5 5
#> GSN GSTP1 GZMB H2AFY H3F3B
#> 3 5 3 5 5
#> HACD4 HCK HIST1H4C HK2 HLA-DPA1
#> 5 5 2 5 3
#> HLA-DPB1 HLA-DQA1 HLA-DRA HLA-DRB1 HLA-DRB6
#> 3 3 3 3 3
#> HMGB2 HNMT HSBP1 HSP90AB1 HSP90AB3P
#> 2 5 5 4 4
#> HSPA1A ID1 IER2 IER3 IER5
#> 5 5 5 5 5
#> IFI30 IFITM3 IFNGR2 IGSF6 IL13RA1
#> 5 5 5 5 5
#> IL1B IL1RN IQGAP1 IRAK3 IRF2BP2
#> 5 5 5 5 5
#> IRF7 IRF8 IRS2 ITGAM ITM2B
#> 3 3 5 5 5
#> ITM2C IVNS1ABP JAML JUN JUNB
#> 3 5 5 5 5
#> JUND KCNE3 KCTD12 KIAA0930 KIF23
#> 5 5 5 5 2
#> KLF10 KLF4 KLF6 KNSTRN LAMP5
#> 5 5 5 2 3
#> LAMTOR1 LAMTOR4 LCP1 LGALS1 LGALS2
#> 5 5 5 5 5
#> LGALS3 LGMN LILRA2 LILRA4 LILRA5
#> 5 3 5 3 5
#> LILRB2 LILRB3 LILRB4 LINC00877 LINC00996
#> 5 5 3 5 3
#> LMO2 LRP1 LRRC26 LRRC36 LRRK2
#> 5 5 3 3 5
#> LSM6 LST1 LTA4H LY86 LY96
#> 5 5 5 5 5
#> LYST LYZ MAFB MAP1A MAPKAPK2
#> 5 5 5 3 3
#> MARCKS MCL1 MEGF9 METTL9 MFSD1
#> 5 5 5 5 5
#> MGST1 MGST2 MIDN MNDA MPEG1
#> 5 3 5 5 5
#> MS4A6A MS4A7 MSRB1 MYADM MYBL2
#> 5 5 5 5 3
#> MYL6 MZB1 NAMPT NAPRT NAPSB
#> 3 3 5 5 3
#> NCF2 NDUFA11 NDUFB1 NDUFS6 NFAM1
#> 5 5 5 5 5
#> NFKBIA NFKBIZ NINJ1 NLRP3 NOTCH2
#> 5 5 5 5 5
#> NPC2 NR4A2 NRG1 NT5DC2 NUDT16
#> 5 5 5 3 5
#> NUP214 NUSAP1 OAZ1 ODF3B OGFRL1
#> 5 2 5 5 5
#> OSBPL8 OSCAR OTUD1 P2RY13 PARK7
#> 5 5 5 5 3
#> PCBP1 PCNA PFDN5 PFN1 PGD
#> 5 1 5 2 5
#> PGLS PHEX PICALM PID1 PILRA
#> 5 3 5 5 5
#> PLA2G7 PLAC8 PLAUR PLBD1 PLD4
#> 5 3 5 5 3
#> PLEK PLP2 PLSCR1 PLXDC2 PLXNB2
#> 3 3 5 5 5
#> PNRC1 POLE4 POLR2L POMP PPP1R14B
#> 5 5 5 5 3
#> PPP1R15A PPT1 PRAM1 PRDX3 PRELID1
#> 5 5 5 5 5
#> PROC PRR11 PSAP PTAFR PTGDS
#> 3 2 5 5 3
#> PTGS2 PTPRE PTPRS PTTG1 PYCARD
#> 5 5 3 2 5
#> PYGL QKI QPCT RAB11FIP1 RAB31
#> 5 5 5 5 5
#> RAB32 RAB3D RAC1 RACGAP1 RASSF2
#> 5 5 5 2 5
#> RBM47 RBP7 RBX1 RETN RGS2
#> 5 5 5 5 5
#> RHOA RHOB RHOG RIPK2 RN7SL368P
#> 5 5 5 5 5
#> RNA5SP151 RNASE2 RNASE6 RNF130 RNU4ATAC
#> 5 5 3 5 5
#> RNVU1-19 RP11-1143G9.4 RP11-160E2.6 RP11-24F11.2 RP11-363K21.1
#> 5 5 5 3 5
#> RP11-40C6.2 RP11-522I20.3 RP11-71G12.1 RP11-73G16.2 RP4-647C14.2
#> 4 2 3 3 3
#> RP5-1121H13.3 RPL13A RPL23A RPL27A RPL3
#> 2 4 4 4 4
#> RPL31 RPL4 RPL5 RPLP2 RPS12
#> 4 4 4 4 4
#> RPS18 RPS2 RPS23 RPS24 RPS27
#> 4 4 4 5 4
#> RPS27A RPS29 RPS3 RPS5 RPS6
#> 4 4 4 4 4
#> RPSA RRM2 RTN1 RTN3 RTN4
#> 4 2 5 5 5
#> RXRA S100A10 S100A11 S100A12 S100A4
#> 5 5 5 5 5
#> S100A6 S100A8 S100A9 SAMHD1 SAT1
#> 5 5 5 5 5
#> SCAMP5 SCN9A SCT SDCBP SEC11A
#> 3 3 3 5 5
#> SEC61B SEC61G SELENOS SEMA4A SERF2
#> 3 3 3 5 5
#> SERPINA1 SERPINF1 SGK1 SH3BGRL SH3BGRL3
#> 5 3 5 5 3
#> SIRPA SIRPB1 SLC11A1 SLC15A3 SLC16A3
#> 5 5 5 5 5
#> SLC31A2 SLC7A7 SLC8A1 SMC2 SMCO4
#> 5 5 5 2 5
#> SMIM25 SMIM5 SMPD3 SNORD3B-2 SOD2
#> 5 3 3 5 5
#> SORT1 SPCS1 SPDL1 SPI1 SRGN
#> 5 3 2 5 5
#> STAB1 STMN1 STX11 STXBP2 SULF2
#> 5 2 5 5 3
#> SULT1A1 SYCP2L TAGLN2 TALDO1 TCF4
#> 5 3 3 5 3
#> TET2 TFEC TGFBI TIMM8B TIMP1
#> 5 5 5 5 5
#> TIMP2 TKT TLR2 TLR4 TLR8
#> 5 5 5 5 5
#> TMEM107 TMEM167A TMEM170B TMEM8B TNFAIP2
#> 5 5 5 3 5
#> TNFRSF1B TNFRSF21 TNFSF10 TNFSF13B TOP2A
#> 5 3 5 5 2
#> TOX2 TPI1 TPM2 TPP1 TPX2
#> 2 5 3 5 2
#> TRAIP TREM1 TRIB1 TSPO TTI2
#> 2 5 5 5 2
#> TUBA1A TUBA1B TUBB TXN TYMP
#> 5 2 2 3 5
#> TYMS TYROBP UBE2C UGCG UQCR11
#> 2 5 2 3 5
#> VAMP8 VCAN VIM VSIR WARS
#> 3 5 5 5 5
#> YBX3 YPEL5 YWHAE ZEB2 ZFAND5
#> 5 5 5 5 5
#> ZFP36 ZFP36L1 ZNF385A ZNF467 ZWINT
#> 5 5 3 5 2
scdb_add_gset(gset_nm, gset)Note that we constructed a membership vector with genes as names and the number of the top correlated anchor gene as the value. This is optional, we could have created a gene set with all genes in the same set (e.g. 1). It is important to add the gene set to the database for the next steps. We use gset_nm as the gene set id.
Fine clustering and filtering of the gene set
To further examine the gene set we got, we'll cluster the genes. First, we'll create a sub-matrix from the umi matrix that will contain only the genes in the gene set:
sub_mat_id = paste("test", gset_nm, sep="_")
mcell_mat_ignore_genes(new_mat_id = sub_mat_id, mat_id = "test", ig_genes = names(foc_genes), reverse = T)Now we'll cluster the genes on the downsampled sub-matrix, using hierarchical clustering and cutting the tree to 20 clusters:
mcell_gset_split_by_dsmat(gset_id = gset_nm, mat_id = sub_mat_id, K = 20)
#> will downsample the matrix, N= 750The number of clusters should be proportional to the number of genes we cluster, as we want to get homogeneous clusters of coodinated genes.
The cluster membership information is updated in the gene set. Let's re-load it and have a look:
gset = scdb_gset(gset_nm)
print(gset)
#> An object of class "tgGeneSets"
#> Slot "description":
#> [1] "Cell cycle and stress correlated genes hc K=20"
#>
#> Slot "set_names":
#> [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
#>
#> Slot "gene_set":
#> AC023590.1 ACTB ACTG1 AGTRAP AHNAK
#> 1 2 3 4 2
#> AHR AIF1 ALDH1A1 ALDH2 ALDH3B1
#> 5 6 7 8 7
#> ALDOA ALOX5AP ANKRD28 ANPEP ANXA1
#> 2 9 5 7 2
#> ANXA2 ANXA5 AOAH AP1S2 AP2S1
#> 2 2 10 4 6
#> APLP2 APP ARHGEF40 ARL5B ARL6IP1P2
#> 4 11 7 5 12
#> ARRDC4 ASAH1 ASF1B ASGR1 ASIP
#> 7 13 14 4 11
#> ATF3 ATG3 ATOX1 ATP2B1-AS1 ATP5E
#> 10 15 5 10 5
#> ATP5G2 ATP5I ATP6V0B ATP6V0D1 ATP6V0E1
#> 5 5 6 7 16
#> ATP6V1A ATP6V1B2 ATP6V1F BACH1 BCAT1
#> 7 15 7 15 12
#> BCL2A1 BIRC5 BLOC1S1 BLVRB BNIP3L
#> 15 14 5 4 5
#> BRI3 BST1 C10orf11 C12orf75 C14orf2
#> 15 4 7 9 7
#> C19orf38 C19orf47 C1orf162 C20orf27 C4orf48
#> 13 12 15 15 7
#> C5AR1 C7orf73 C9orf72 CALM2 CAMK1
#> 13 5 15 15 15
#> CAPG CAPZA2 CARD16 CARS2 CASP1
#> 8 15 15 7 15
#> CATSPER1 CCDC189 CCDC50 CCDC71L CCDC88A
#> 10 9 11 12 7
#> CCL3 CCL3L3 CCNA2 CCNL1 CCNY
#> 10 10 14 17 5
#> CCR1 CCR9 CD14 CD163 CD300E
#> 15 12 4 10 15
#> CD300LF CD302 CD33 CD36 CD4
#> 15 10 4 4 15
#> CD44 CD63 CD70 CD83 CD86
#> 16 8 12 15 15
#> CD93 CDA CDC42 CDCA8 CDK1
#> 10 13 16 14 14
#> CDK2 CDKN2C CDKN3 CDR2 CEBPB
#> 12 12 14 16 13
#> CEBPD CECR1 CENPE CENPF CENPN
#> 6 10 14 14 12
#> CENPP CENPU CFD CFP CHMP1B
#> 14 14 6 6 16
#> CHP1 CIB2 CKAP4 CKLF CLEC10A
#> 7 9 7 2 18
#> CLEC12A CLEC4A CLEC4C CLEC4E CLEC7A
#> 13 4 11 4 6
#> CMTM6 CNPY3 COMT CORO1A COTL1
#> 15 8 5 3 2
#> COX6B1 COX7B CPPED1 CPVL CREB5
#> 5 5 15 6 7
#> CREG1 CRTAP CRYM CSF3R CST3
#> 7 7 1 4 6
#> CSTA CTB-61M7.2 CTC-510F12.4 CTD-3252C9.4 CTSB
#> 6 7 7 10 8
#> CTSC CTSD CTSH CTSS CTSZ
#> 9 4 19 6 6
#> CXCL2 CXCL8 CYBA CYBB CYP1B1
#> 7 10 6 6 7
#> DAZAP2 DERL3 DMXL2 DNASE1L3 DOK3
#> 5 11 15 11 15
#> DPM2 DPYD DPYSL2 DSTNP2 DUSP1
#> 12 5 10 12 17
#> DUSP6 DYNLL1 EFCAB11 EGR1 EHBP1L1
#> 13 5 12 17 7
#> EIF1 EIF4EBP1 ELOB ENHO ENTPD1
#> 17 15 5 18 15
#> EREG EVI2A EVI2B FAM120A FAM198B
#> 7 16 15 5 7
#> FAM45A FANCI FCAR FCER1A FCER1G
#> 7 12 7 18 6
#> FCGR1A FCGR2A FCGRT FCN1 FES
#> 7 15 6 6 8
#> FGD4 FGL2 FKBP1A FKBP2 FLT3
#> 15 6 15 12 9
#> FOLR3 FOS FOSB FOSL2 FPR1
#> 5 17 17 7 4
#> FTH1 FTL FTLP3 G0S2 GABARAPL1
#> 6 6 13 7 7
#> GAPDH GAS6 GAS7 GCA GLIPR1
#> 2 1 5 4 8
#> GLRX GLUL GMFG GNAQ GNS
#> 7 5 16 5 15
#> GPBAR1 GPCPD1 GPX1 GRINA GRN
#> 13 7 4 15 4
#> GSN GSTP1 GZMB H2AFY H3F3B
#> 18 6 11 8 17
#> HACD4 HCK HIST1H4C HK2 HLA-DPA1
#> 15 13 3 7 19
#> HLA-DPB1 HLA-DQA1 HLA-DRA HLA-DRB1 HLA-DRB6
#> 19 19 19 19 19
#> HMGB2 HNMT HSBP1 HSP90AB1 HSP90AB3P
#> 17 10 6 20 12
#> HSPA1A ID1 IER2 IER3 IER5
#> 5 10 17 15 7
#> IFI30 IFITM3 IFNGR2 IGSF6 IL13RA1
#> 13 13 15 6 15
#> IL1B IL1RN IQGAP1 IRAK3 IRF2BP2
#> 10 7 15 7 5
#> IRF7 IRF8 IRS2 ITGAM ITM2B
#> 11 11 16 7 16
#> ITM2C IVNS1ABP JAML JUN JUNB
#> 11 16 8 17 17
#> JUND KCNE3 KCTD12 KIAA0930 KIF23
#> 10 7 10 15 12
#> KLF10 KLF4 KLF6 KNSTRN LAMP5
#> 10 10 17 12 1
#> LAMTOR1 LAMTOR4 LCP1 LGALS1 LGALS2
#> 15 5 5 6 6
#> LGALS3 LGMN LILRA2 LILRA4 LILRA5
#> 6 9 15 11 13
#> LILRB2 LILRB3 LILRB4 LINC00877 LINC00996
#> 13 13 1 15 11
#> LMO2 LRP1 LRRC26 LRRC36 LRRK2
#> 15 15 1 12 15
#> LSM6 LST1 LTA4H LY86 LY96
#> 5 6 15 19 10
#> LYST LYZ MAFB MAP1A MAPKAPK2
#> 13 6 13 1 1
#> MARCKS MCL1 MEGF9 METTL9 MFSD1
#> 15 5 7 7 7
#> MGST1 MGST2 MIDN MNDA MPEG1
#> 4 8 7 6 6
#> MS4A6A MS4A7 MSRB1 MYADM MYBL2
#> 4 13 7 16 1
#> MYL6 MZB1 NAMPT NAPRT NAPSB
#> 2 11 13 5 11
#> NCF2 NDUFA11 NDUFB1 NDUFS6 NFAM1
#> 13 5 5 5 15
#> NFKBIA NFKBIZ NINJ1 NLRP3 NOTCH2
#> 10 15 15 7 15
#> NPC2 NR4A2 NRG1 NT5DC2 NUDT16
#> 6 17 7 12 15
#> NUP214 NUSAP1 OAZ1 ODF3B OGFRL1
#> 4 14 6 13 10
#> OSBPL8 OSCAR OTUD1 P2RY13 PARK7
#> 12 15 7 5 3
#> PCBP1 PCNA PFDN5 PFN1 PGD
#> 5 12 16 2 7
#> PGLS PHEX PICALM PID1 PILRA
#> 15 1 7 10 13
#> PLA2G7 PLAC8 PLAUR PLBD1 PLD4
#> 7 3 13 4 11
#> PLEK PLP2 PLSCR1 PLXDC2 PLXNB2
#> 8 9 15 10 15
#> PNRC1 POLE4 POLR2L POMP PPP1R14B
#> 17 5 7 5 11
#> PPP1R15A PPT1 PRAM1 PRDX3 PRELID1
#> 17 15 13 15 15
#> PROC PRR11 PSAP PTAFR PTGDS
#> 11 12 6 7 1
#> PTGS2 PTPRE PTPRS PTTG1 PYCARD
#> 7 10 9 14 6
#> PYGL QKI QPCT RAB11FIP1 RAB31
#> 7 7 4 7 7
#> RAB32 RAB3D RAC1 RACGAP1 RASSF2
#> 4 7 6 12 7
#> RBM47 RBP7 RBX1 RETN RGS2
#> 7 4 5 7 6
#> RHOA RHOB RHOG RIPK2 RN7SL368P
#> 5 13 2 7 7
#> RNA5SP151 RNASE2 RNASE6 RNF130 RNU4ATAC
#> 7 7 8 6 7
#> RNVU1-19 RP11-1143G9.4 RP11-160E2.6 RP11-24F11.2 RP11-363K21.1
#> 16 4 7 9 15
#> RP11-40C6.2 RP11-522I20.3 RP11-71G12.1 RP11-73G16.2 RP4-647C14.2
#> 20 12 1 11 1
#> RP5-1121H13.3 RPL13A RPL23A RPL27A RPL3
#> 16 20 20 20 20
#> RPL31 RPL4 RPL5 RPLP2 RPS12
#> 20 20 20 20 20
#> RPS18 RPS2 RPS23 RPS24 RPS27
#> 20 20 20 12 20
#> RPS27A RPS29 RPS3 RPS5 RPS6
#> 20 20 20 20 20
#> RPSA RRM2 RTN1 RTN3 RTN4
#> 20 14 15 5 5
#> RXRA S100A10 S100A11 S100A12 S100A4
#> 15 2 6 4 6
#> S100A6 S100A8 S100A9 SAMHD1 SAT1
#> 6 4 4 5 6
#> SCAMP5 SCN9A SCT SDCBP SEC11A
#> 1 9 11 15 5
#> SEC61B SEC61G SELENOS SEMA4A SERF2
#> 9 9 9 15 5
#> SERPINA1 SERPINF1 SGK1 SH3BGRL SH3BGRL3
#> 6 11 10 15 2
#> SIRPA SIRPB1 SLC11A1 SLC15A3 SLC16A3
#> 15 7 13 15 7
#> SLC31A2 SLC7A7 SLC8A1 SMC2 SMCO4
#> 15 13 15 12 15
#> SMIM25 SMIM5 SMPD3 SNORD3B-2 SOD2
#> 13 11 1 15 7
#> SORT1 SPCS1 SPDL1 SPI1 SRGN
#> 15 9 12 6 6
#> STAB1 STMN1 STX11 STXBP2 SULF2
#> 7 3 15 6 7
#> SULT1A1 SYCP2L TAGLN2 TALDO1 TCF4
#> 15 12 9 8 11
#> TET2 TFEC TGFBI TIMM8B TIMP1
#> 5 7 8 5 2
#> TIMP2 TKT TLR2 TLR4 TLR8
#> 7 13 15 7 7
#> TMEM107 TMEM167A TMEM170B TMEM8B TNFAIP2
#> 5 7 7 9 10
#> TNFRSF1B TNFRSF21 TNFSF10 TNFSF13B TOP2A
#> 15 11 15 4 14
#> TOX2 TPI1 TPM2 TPP1 TPX2
#> 1 2 11 8 14
#> TRAIP TREM1 TRIB1 TSPO TTI2
#> 12 7 7 6 12
#> TUBA1A TUBA1B TUBB TXN TYMP
#> 17 3 3 9 6
#> TYMS TYROBP UBE2C UGCG UQCR11
#> 14 6 14 11 16
#> VAMP8 VCAN VIM VSIR WARS
#> 5 4 2 2 13
#> YBX3 YPEL5 YWHAE ZEB2 ZFAND5
#> 10 17 8 6 15
#> ZFP36 ZFP36L1 ZNF385A ZNF467 ZWINT
#> 17 5 15 7 14However, it is much more convenient to look at the correlation heatmaps of these clusters, so let's generate those:
mcell_plot_gset_cor_mats(gset_id = gset_nm, scmat_id = sub_mat_id)
#> will downsample the matrix, N= 750The plots are generated under a subdirectory of the figures directory named gset_nm.gset_cors. Common practice is to examine them and select the clusters that we want to keep. In our example, In this case we would probably keep clusters 3, 5, 10, 14, 16 and 17 in the gene set: 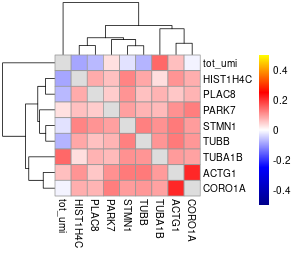
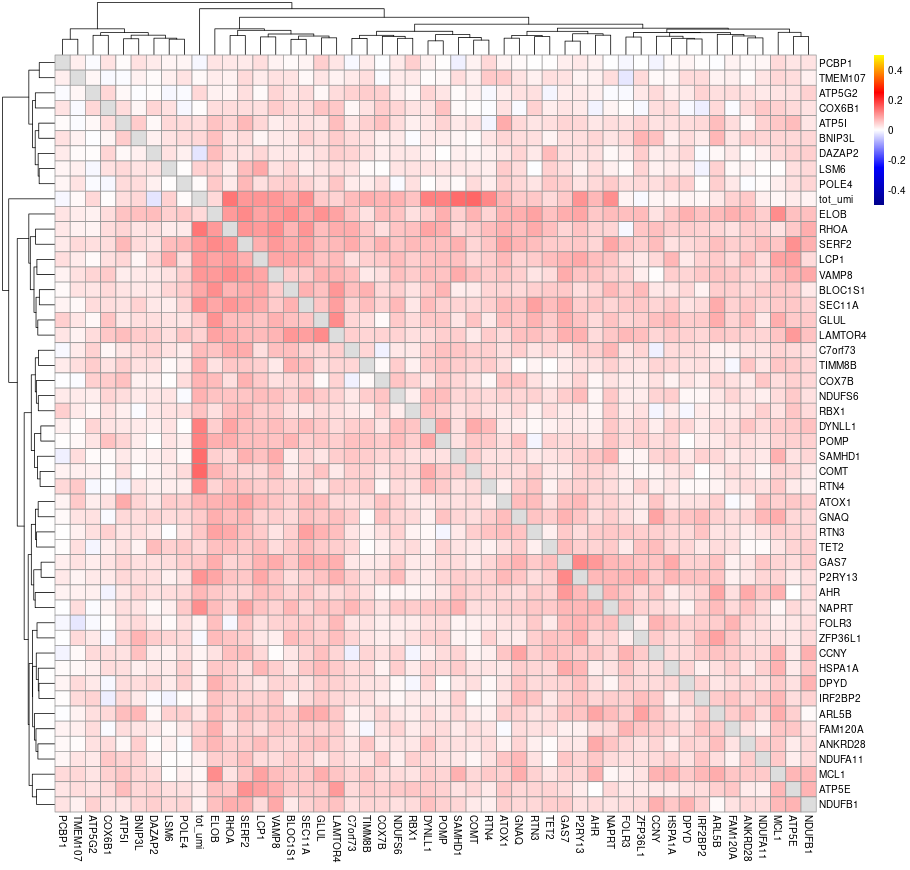
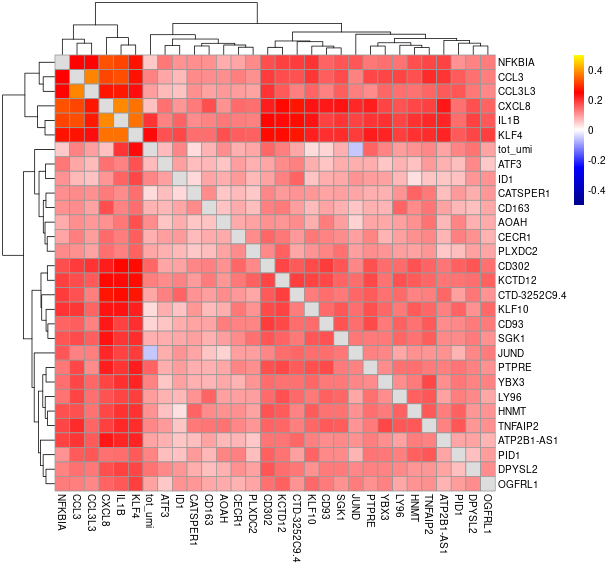
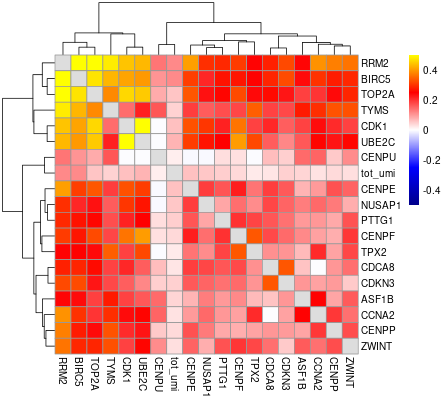
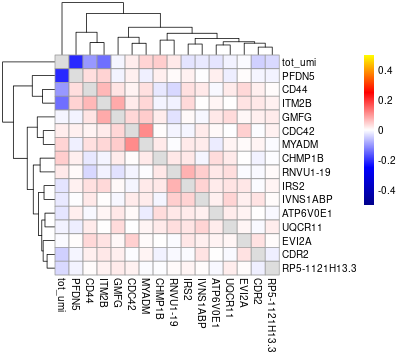
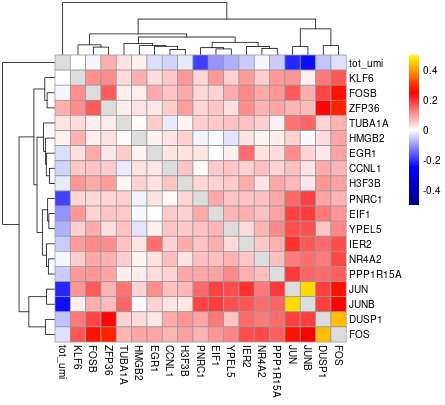
The rest of the clusters contain mostly genes that are correlated to cell size but not related to the cell cycle.
To create the final filtered gene set, we'll keep only the clusters we've selected in the gene set, and we'll save the filtered gene set in the database:
mcell_gset_remove_clusts(gset_id = gset_nm, filt_clusts = c(3, 5, 10, 14, 16, 17), new_id = paste0(gset_nm, "_filtered"), reverse=T)
lateral_gset_id = paste0(gset_nm, "_filtered")
lateral_gset = scdb_gset(lateral_gset_id)
print(lateral_gset)
#> An object of class "tgGeneSets"
#> Slot "description":
#> [1] "Cell cycle and stress correlated genes hc K=20 cl_filt"
#>
#> Slot "set_names":
#> [1] 3 5 10 14 16 17
#>
#> Slot "gene_set":
#> ACTG1 AHR ANKRD28 AOAH ARL5B
#> 3 5 5 10 5
#> ASF1B ATF3 ATOX1 ATP2B1-AS1 ATP5E
#> 14 10 5 10 5
#> ATP5G2 ATP5I ATP6V0E1 BIRC5 BLOC1S1
#> 5 5 16 14 5
#> BNIP3L C7orf73 CATSPER1 CCL3 CCL3L3
#> 5 5 10 10 10
#> CCNA2 CCNL1 CCNY CD163 CD302
#> 14 17 5 10 10
#> CD44 CD93 CDC42 CDCA8 CDK1
#> 16 10 16 14 14
#> CDKN3 CDR2 CECR1 CENPE CENPF
#> 14 16 10 14 14
#> CENPP CENPU CHMP1B COMT CORO1A
#> 14 14 16 5 3
#> COX6B1 COX7B CTD-3252C9.4 CXCL8 DAZAP2
#> 5 5 10 10 5
#> DPYD DPYSL2 DUSP1 DYNLL1 EGR1
#> 5 10 17 5 17
#> EIF1 ELOB EVI2A FAM120A FOLR3
#> 17 5 16 5 5
#> FOS FOSB GAS7 GLUL GMFG
#> 17 17 5 5 16
#> GNAQ H3F3B HIST1H4C HMGB2 HNMT
#> 5 17 3 17 10
#> HSPA1A ID1 IER2 IL1B IRF2BP2
#> 5 10 17 10 5
#> IRS2 ITM2B IVNS1ABP JUN JUNB
#> 16 16 16 17 17
#> JUND KCTD12 KLF10 KLF4 KLF6
#> 10 10 10 10 17
#> LAMTOR4 LCP1 LSM6 LY96 MCL1
#> 5 5 5 10 5
#> MYADM NAPRT NDUFA11 NDUFB1 NDUFS6
#> 16 5 5 5 5
#> NFKBIA NR4A2 NUSAP1 OGFRL1 P2RY13
#> 10 17 14 10 5
#> PARK7 PCBP1 PFDN5 PID1 PLAC8
#> 3 5 16 10 3
#> PLXDC2 PNRC1 POLE4 POMP PPP1R15A
#> 10 17 5 5 17
#> PTPRE PTTG1 RBX1 RHOA RNVU1-19
#> 10 14 5 5 16
#> RP5-1121H13.3 RRM2 RTN3 RTN4 SAMHD1
#> 16 14 5 5 5
#> SEC11A SERF2 SGK1 STMN1 TET2
#> 5 5 10 3 5
#> TIMM8B TMEM107 TNFAIP2 TOP2A TPX2
#> 5 5 10 14 14
#> TUBA1A TUBA1B TUBB TYMS UBE2C
#> 17 3 3 14 14
#> UQCR11 VAMP8 YBX3 YPEL5 ZFP36
#> 16 5 10 17 17
#> ZFP36L1 ZWINT
#> 5 14Generating metacells while blacklisting our lateral gene set
This section will follow the exact same steps performed in the Running metacell analysis: guided tutorial on 8K PBMCs vignette in order to generate the K-nn graph, the co-clustering matrix and the metacells, with the small change of disabling the lateral genes from affecting the process.
We start by removing the lateral genes from the list of feature genes:
marker_gset = scdb_gset("test_feats")
marker_gset = gset_new_restrict_gset(gset = marker_gset, filt_gset = lateral_gset, inverse = T, desc = "cgraph gene markers w/o lateral genes")
scdb_add_gset("test_feats_filtered", marker_gset)We'll now generate the graph using the filtered list of feature genes:
mcell_add_cgraph_from_mat_bknn(mat_id="test",
gset_id = "test_feats_filtered",
graph_id="test_graph_filtered",
K=100,
dsamp=T)
#> will downsample the matrix, N= 1877
#> will build balanced knn graph on 8276 cells and 851 genes, this can be a bit heavy for >20,000 cells
#> sim graph is missing 36 nodes, out of 8276And now simply following the steps of the Running metacell analysis: guided tutorial on 8K PBMCs vignette from the K-nn graph until the colored metacell object:
mcell_coclust_from_graph_resamp(
coc_id="test_coc500_filtered",
graph_id="test_graph_filtered",
min_mc_size=20,
p_resamp=0.75, n_resamp=500)
#> running bootstrap to generate cocluster
#> done resampling
mcell_mc_from_coclust_balanced(
coc_id="test_coc500_filtered",
mat_id= "test",
mc_id= "test_filtered_mc",
K=30, min_mc_size=30, alpha=2)
#> filtered 5027554 left with 742806 based on co-cluster imbalance
#> building metacell object, #mc 75
#> add batch counts
#> compute footprints
#> compute absolute ps
#> compute coverage ps
#> reordering metacells by hclust and most variable two markers
#> reorder on CD74 vs LEF1
mcell_mc_split_filt(new_mc_id="test_filtered_mc_f",
mc_id="test_filtered_mc",
mat_id="test",
T_lfc=3, plot_mats=F)
#> starting split outliers
#> add batch counts
#> compute footprints
#> compute absolute ps
#> compute coverage ps
marks_colors = read.table(system.file("extdata", "pbmc_mc_colorize.txt", package="metacell"), sep="\t", h=T, stringsAsFactors=F)
mc_colorize("test_filtered_mc_f", marker_colors=marks_colors)It is convenient not to restrict the selection of genes that will be displayed in the heatmap of genes and metacells, but to mark the lateral genes that did not affect the partitioning of cells to metacells:
mcell_gset_from_mc_markers(gset_id="test_filtered_markers", mc_id="test_filtered_mc_f", blacklist_gset_id=lateral_gset_id)
mcell_gset_from_mc_markers(gset_id="test_filtered_markers_lat", mc_id="test_filtered_mc_f", filt_gset_id=lateral_gset_id)
mcell_mc_plot_marks(mc_id="test_filtered_mc_f", gset_id="test_filtered_markers", mat_id="test", lateral_gset="test_filtered_markers_lat")
#> setting fig h to 1000 md levels 0 num of marks 47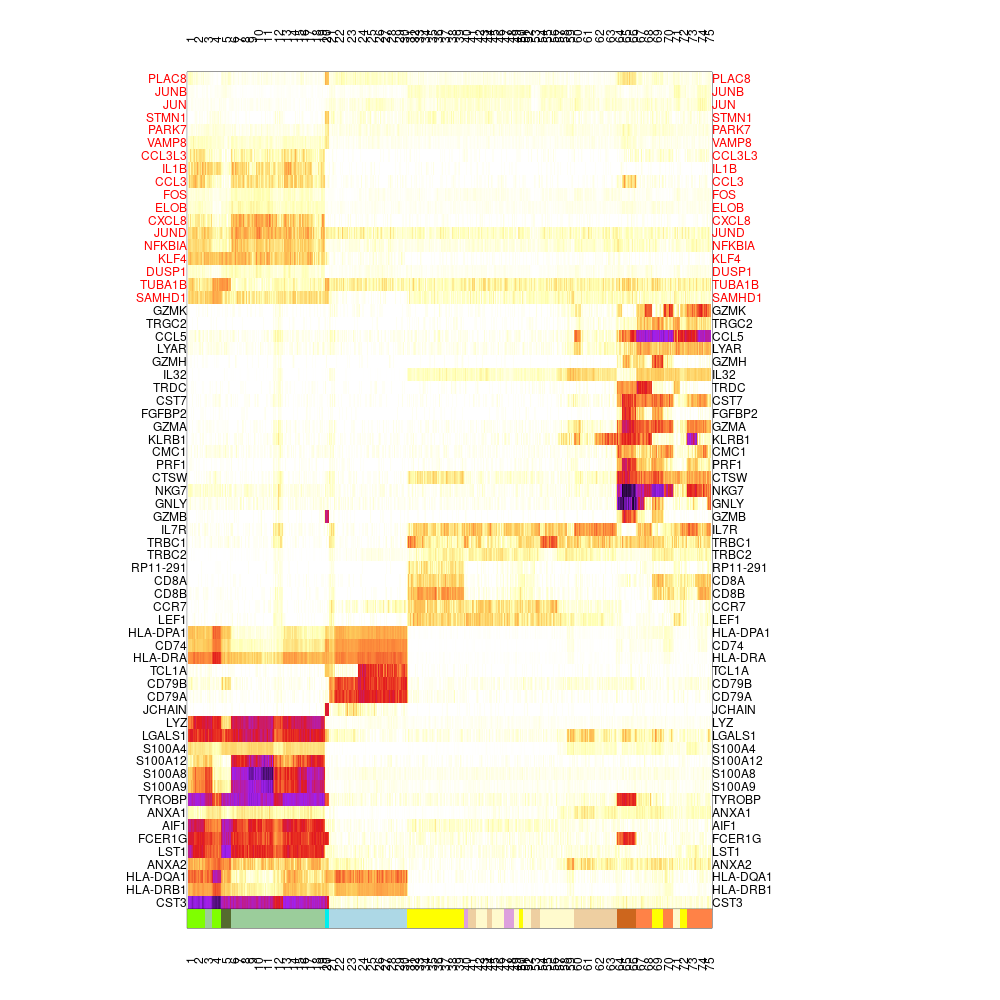 The marker genes in the test_filtered_markers gene set do not contain any lateral gene (shown in black in the heatmap), and the test_filtered_markers_lat gene set contain the lateral genes that were selected as markers (shown in red).
The marker genes in the test_filtered_markers gene set do not contain any lateral gene (shown in black in the heatmap), and the test_filtered_markers_lat gene set contain the lateral genes that were selected as markers (shown in red).